I am working on a genome assembly- this is a human sized vertebrate genome (Peromyscus eremicus). I work on this species in the context of adaptation to deserts (e.g. extreme heat and aridity), and in the future on its behavior. Aside from the biological goals, I’m interested in showing that large genome assembly projects don’t need huge consortiums or massive budgets. To meet the needs of many projects (e.g. genome for use in studies of (comparative) functional biology), these assemblies can be done with modest budgets by a single lab. This is a work in progress, so we’ll see if I’m right..
The data consist of 5 PE lanes and 1 lane from 2 MP libraries (5kb and 7kb insert) of read length 150nt. All of these are from rapid runs on a HiSeq 2500. In addition to this I have 3x coverage PacBio data fom the new P5-C3 chemistries (read N50=8800nt, max=38kb). As you can see, this is very modest. I’ll do some more sequencing, but we’ll see how far this gets me.
FYI, I’m using a CRAY supercomputer for unitig construction. I’ve split up my input files into 145 equally sized chunks, ran with 1536 nodes — ABySS absolutely flies like this. Unitig construction done in about 75 minutes!!! I use XSEDE Blacklight for the contig construction and scaffolding.
So the question for today is of parameter optimization. I’m using ABySS, and in the process of optimizing k at the unitig stage. The issue I have, and everybody doing genome assembly has, is what parameter to optimize. See the table below, red number are the ‘optimum’ for a given parameter:
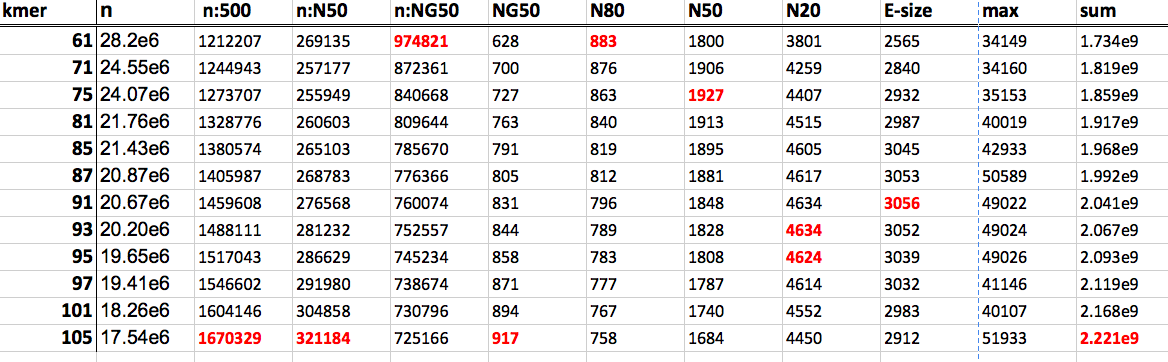
Optimize contiguity is the goal, but unfortunately (cue Assemblathon 2), different measures of contiguity suggest different values of k. NG50 suggests k=105, N50 suggests k=75, and finally E-size (from GAGE paper) suggests k=91… Further, given all of the contiguity stats are relatively similar, at least between the larger values for k, maybe I should decide based on the size of the assembly. For ABySS, if a given kmer (or contig) is absent from the unitig assembly, it’s absent from all other stages, so maybe maximizing the size of the assembly (within reason) is something to consider. You can always get rid of contigs later on in the process, I suppose..
So, some outstanding questions:
- What k is best? I think I’ll go with 105, thought the kmers 91,93,95 look good, too.
- How is the k that maximizes unitig construction related to the k that maximizes contig assembly (and later, scaffolding)? Are these going to be different? If so, how different?